After installing Lapack and Blas, I tried to install lapack++ with failure from http://switch.dl.sourceforge.net/sourceforge/lapackpp/lapackpp-2.5.2.tar.gz
I type ./configure
while I have libblas.a liblapack.a librefblas.a libtmglib.a in my use/local/lin folder.... Can any one help?
- 2 Contributors
- forum4 Replies
- 506 Views
- 3 Hours Discussion Span
- commentLatest PostLatest Postby cannon_1
Recommended Answers
- A number of different LAPACK library setups can be used, including optimized LAPACK libraries such as ATLAS, MKL or the Accelerate/vecLib framework on OS X. FORTRAN ABI mismatch ¶ The two most popular open source fortran compilers are g77 and gfortran.
- On Ubuntu, such libraries can be made available by installing liblapack-dev. Other Linux distributions typically also offer a way to install Blas and Lapack via its package manager. On Mac OS X, Ipopt's build system should pick up the Blas and Lapack from the Accelerate framework.
Are you sure you have compatible versions of Blas and Lapack installed? Also, what is the output of ../configure when you run it in the extracted directory for lapack++?
Jump to PostAll 4 Replies
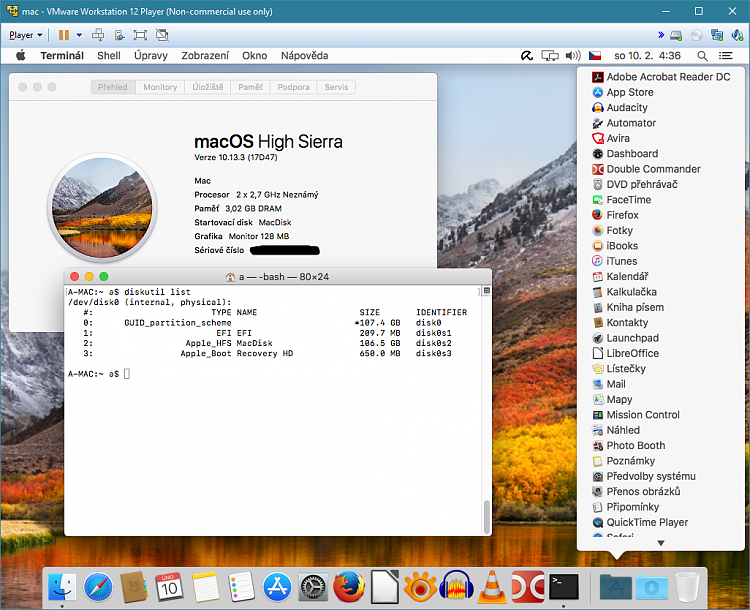
Are you sure you have compatible versions of Blas and Lapack installed? Also, what is the output of ../configure when you run it in the extracted directory for lapack++?
Installing Plumed
Plumed is an exiting 'new' rare-event code that is an open source library for free energy calculations in molecular systems which works together with some of the most popular molecular dynamics engines. Free energy calculations can be performed as a function of many order parameters with a particular focus on biological problems, using state of the art methods such as metadynamics, umbrella sampling and Jarzynski-equation based steered MD. The software, written in C++, can be easily interfaced with both fortran and C/C++ codes.
However with the advent of PLUMED2 – v2.0.1 in late 2013, the code had major changes for the user interface. Over the next few posts I will provide Installations and basic user guides for the new version of Plumed.
1. Installing Libraries
If the BLAS and LAPACK shared and/or static libraries are installed, there should be no further problems following the installation instructions below. Mac OS X A binary can be built for Mac using Apple's optimized versions of the BLAS and LAPACK libraries, which are contained in the 'vecLib' sub-framework, part of the 'Accelerate' framework.
Before you install Plumed you need to install BLAS and LAPAC libraries. You can download them together at – http://www.netlib.org/lapack/#_software. Download 'lapack-3.5.0'. Put them in the relevant directory and unzip the folder.
Installing BLAS
Step 1 – cd /lapack-3.5.0/BLAS/SRC
Step 2 – Compile all of the .f files to produce .o files
g77 -c -O3 *.f
Step 3 – Combine the .o files into a library
ar rv libblas.a *.o
Step 4 – Switch to root and install
cp libblas.a /usr/local/lib
Installing LAPACK
Step 1 – cd /lapack-3.5.0/
Step 2 – You can either use pre-compiled installation. Copy the appropriate make.inc.XXX file from the INSTALL directory into make.inc in the main directory.
Example: cp INSTALL/make.inc.LINUX make.inc
OR
./Cmake
Step 3 – sudo make
Mac Os X 10.11 Download Free
Step 4 – sudo make install
2. Installing Plumed
Installing Lapack On Mac Os X 10 13 Download
Step 1 – Download Plumed from http://plumed.github.io/doc-v2.0/user-doc/html/_installation.html
Step 2 – cd /plumed-2.0.2
Step 3 – ./configure.sh -ldl -L/usr/local/lib/liblapack.a -llapack -D__PLUMED_HAS_LAPACK -L/usr/local/lib/libblas.a -lblas -D__PLUMED_HAS_BLAS SOEXT=dylib
Select – 3> mac.mpi.clang
Step 4 – Remove flag '-lmpi_cxx' from variable DYNAMIC_LIBS in Makefile.conf
Step 5 – source sourceme.sh
Lapack Download Windows
Step 6 – sudo make
Step 7 – sudo make install
To uninstall, remove the following files and directories:
/usr/local/lib/plumed/
/usr/local/include/plumed
/usr/local/bin/plumed
/usr/local/bin/plumed-mklib
/usr/local/bin/plumed-newcv
/usr/local/bin/plumed-patch
/usr/local/lib/libplumed.dylib
/usr/local/lib/libplumedKernel.dylib
3. Installing Gromacs
We are going to install a mpi version of double precision gromacs with plumed.
For more information on custom Gromacs installation – http://dqfnet.ufpe.br/groups/geekstuff/wiki/2ebb2/
Also Plumed has been tested with a few verions of Gromacs. Ofcourse you can try a different version but it is safest to use the one recommended.
Step 1 – Get gromacs-4.6.5. http://www.gromacs.org/Downloads
Step 2 – cp /patches/patch.sh .
-> 2) gromacs-4.6.5
Step 3 – Make a directory to compile Gromacs. Usually this is recommended as you can end up multiple version of Gromacs in the same machine.
mkdir build
cd build
Step 4 – Make a version of Gromacs with the correct ingredients. Like this is a double precision version with MPI.
CMAKE_PREFIX_PATH=/usr/local/include:/usr/local/lib:/opt/local/:/usr/local/bin cmake .. -DGMX_X11=ON -DBUILD_SHARED_LIBS=OFF -DCMAKE_INSTALL_PREFIX=/usr/local/gromacs4.6.5_double/ -DGMX_DOUBLE=ON -DGMX_MPI=ON -DCMAKE_C_COMPILER=gcc -DCMAKE_CXX_COMPILER=g++ -DMX_GSL=ON -DMX_THREAD_MPI=ON -DGMX_CPU_ACCELERATION=SSE2 -DREGRESSIONTEST_DOWNLOAD=ON -DGMX_BUILD_OWN_FFTW=ON
Step 5 – sudo make
Step 6 – sudo make install
4. Installing Gromacs in Linux
Follow the same steps as above.
CMAKE_PREFIX_PATH=/usr/local/include:/usr/local/lib:/opt/local/:/usr/local/bin cmake .. -DGMX_X11=ON -DBUILD_SHARED_LIBS=OFF -DCMAKE_INSTALL_PREFIX=/usr/local/gromacs4.6.5_double/ -DGMX_DOUBLE=ON -DGMX_MPI=ON -DCMAKE_C_COMPILER=gcc -DCMAKE_CXX_COMPILER=g++ -DMX_GSL=ON -DMX_THREAD_MPI=ON -DGMX_CPU_ACCELERATION=SSE2 -DREGRESSIONTEST_DOWNLOAD=ON
5. Citing PLUMED

Are you sure you have compatible versions of Blas and Lapack installed? Also, what is the output of ../configure when you run it in the extracted directory for lapack++?
Installing Plumed
Plumed is an exiting 'new' rare-event code that is an open source library for free energy calculations in molecular systems which works together with some of the most popular molecular dynamics engines. Free energy calculations can be performed as a function of many order parameters with a particular focus on biological problems, using state of the art methods such as metadynamics, umbrella sampling and Jarzynski-equation based steered MD. The software, written in C++, can be easily interfaced with both fortran and C/C++ codes.
However with the advent of PLUMED2 – v2.0.1 in late 2013, the code had major changes for the user interface. Over the next few posts I will provide Installations and basic user guides for the new version of Plumed.
1. Installing Libraries
If the BLAS and LAPACK shared and/or static libraries are installed, there should be no further problems following the installation instructions below. Mac OS X A binary can be built for Mac using Apple's optimized versions of the BLAS and LAPACK libraries, which are contained in the 'vecLib' sub-framework, part of the 'Accelerate' framework.
Before you install Plumed you need to install BLAS and LAPAC libraries. You can download them together at – http://www.netlib.org/lapack/#_software. Download 'lapack-3.5.0'. Put them in the relevant directory and unzip the folder.
Installing BLAS
Step 1 – cd /lapack-3.5.0/BLAS/SRC
Step 2 – Compile all of the .f files to produce .o files
g77 -c -O3 *.f
Step 3 – Combine the .o files into a library
ar rv libblas.a *.o
Step 4 – Switch to root and install
cp libblas.a /usr/local/lib
Installing LAPACK
Step 1 – cd /lapack-3.5.0/
Step 2 – You can either use pre-compiled installation. Copy the appropriate make.inc.XXX file from the INSTALL directory into make.inc in the main directory.
Example: cp INSTALL/make.inc.LINUX make.inc
OR
./Cmake
Step 3 – sudo make
Mac Os X 10.11 Download Free
Step 4 – sudo make install
2. Installing Plumed
Installing Lapack On Mac Os X 10 13 Download
Step 1 – Download Plumed from http://plumed.github.io/doc-v2.0/user-doc/html/_installation.html
Step 2 – cd /plumed-2.0.2
Step 3 – ./configure.sh -ldl -L/usr/local/lib/liblapack.a -llapack -D__PLUMED_HAS_LAPACK -L/usr/local/lib/libblas.a -lblas -D__PLUMED_HAS_BLAS SOEXT=dylib
Select – 3> mac.mpi.clang
Step 4 – Remove flag '-lmpi_cxx' from variable DYNAMIC_LIBS in Makefile.conf
Step 5 – source sourceme.sh
Lapack Download Windows
Step 6 – sudo make
Step 7 – sudo make install
To uninstall, remove the following files and directories:
/usr/local/lib/plumed/
/usr/local/include/plumed
/usr/local/bin/plumed
/usr/local/bin/plumed-mklib
/usr/local/bin/plumed-newcv
/usr/local/bin/plumed-patch
/usr/local/lib/libplumed.dylib
/usr/local/lib/libplumedKernel.dylib
3. Installing Gromacs
We are going to install a mpi version of double precision gromacs with plumed.
For more information on custom Gromacs installation – http://dqfnet.ufpe.br/groups/geekstuff/wiki/2ebb2/
Also Plumed has been tested with a few verions of Gromacs. Ofcourse you can try a different version but it is safest to use the one recommended.
Step 1 – Get gromacs-4.6.5. http://www.gromacs.org/Downloads
Step 2 – cp /patches/patch.sh .
-> 2) gromacs-4.6.5
Step 3 – Make a directory to compile Gromacs. Usually this is recommended as you can end up multiple version of Gromacs in the same machine.
mkdir build
cd build
Step 4 – Make a version of Gromacs with the correct ingredients. Like this is a double precision version with MPI.
CMAKE_PREFIX_PATH=/usr/local/include:/usr/local/lib:/opt/local/:/usr/local/bin cmake .. -DGMX_X11=ON -DBUILD_SHARED_LIBS=OFF -DCMAKE_INSTALL_PREFIX=/usr/local/gromacs4.6.5_double/ -DGMX_DOUBLE=ON -DGMX_MPI=ON -DCMAKE_C_COMPILER=gcc -DCMAKE_CXX_COMPILER=g++ -DMX_GSL=ON -DMX_THREAD_MPI=ON -DGMX_CPU_ACCELERATION=SSE2 -DREGRESSIONTEST_DOWNLOAD=ON -DGMX_BUILD_OWN_FFTW=ON
Step 5 – sudo make
Step 6 – sudo make install
4. Installing Gromacs in Linux
Follow the same steps as above.
CMAKE_PREFIX_PATH=/usr/local/include:/usr/local/lib:/opt/local/:/usr/local/bin cmake .. -DGMX_X11=ON -DBUILD_SHARED_LIBS=OFF -DCMAKE_INSTALL_PREFIX=/usr/local/gromacs4.6.5_double/ -DGMX_DOUBLE=ON -DGMX_MPI=ON -DCMAKE_C_COMPILER=gcc -DCMAKE_CXX_COMPILER=g++ -DMX_GSL=ON -DMX_THREAD_MPI=ON -DGMX_CPU_ACCELERATION=SSE2 -DREGRESSIONTEST_DOWNLOAD=ON
5. Citing PLUMED
Installing Lapack On Mac Os X 10.10
You may wish to cite the following reference if you have utilized PLUMED in your work:
1. M. Bonomi, D. Branduardi, G. Bussi, C. Camilloni, D. Provasi, P. Raiteri, D. Donadio, F. Marinelli, F. Pietrucci, R.A. Broglia and M. Parrinello.
2. PLUMED: a portable plugin for free-energy calculations with molecular dynamics, Comp. Phys. Comm. 2009 vol. 180 (10) pp. 1961-1972.
